Fixing Landmarks
Image of thing to try:
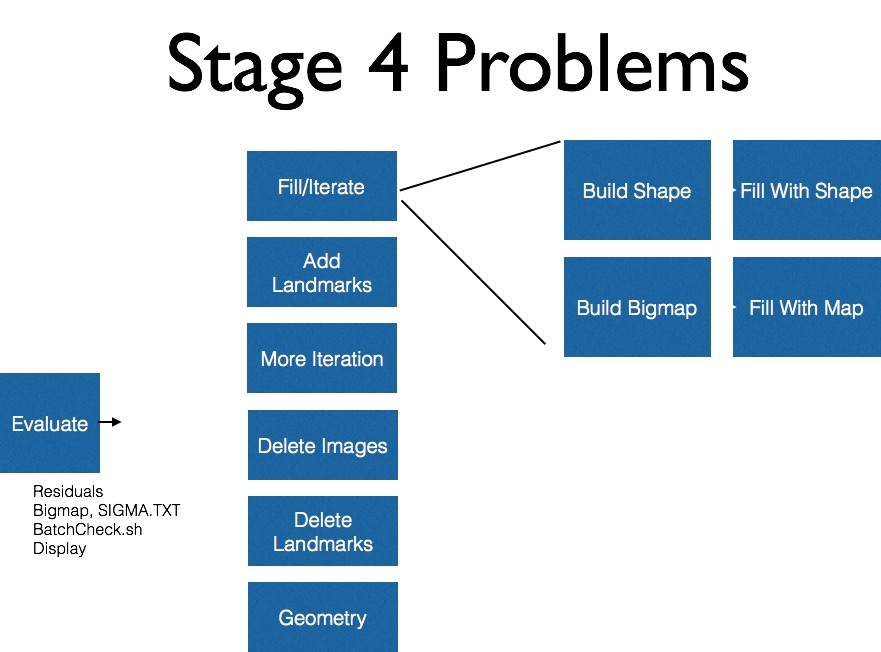
When to Fix Landmarks
At the end of each tiling or iteration, a form of find_nofit (T or P) is used to check for bad landmarks.
A common find_nofit output will look soemthing like this:
EQ0004: cat 004.OOT EP0002: cat 005.OOT EP0003: cat 006.OOT EO0003: cat 007.OOT EN0003: * cat 008.OOT EN0004: * cat 009.OOT EM0003: . cat 010.OOT EM0004: * cat 011.OOT 132 deleted ! cat 132.OOTThe
The Four main types of find_nofit outputs are:
- ! - the landmark was deleted
- . - the landmark has overlaps
- (space) - the landmark has minor error (don't worry about it).
- * - the landmark has a major error and must be fixed.
All errors are worth recording, but the only output from find_nofit that warrants serious concern is *. Whenever a * is present in the find_nofit output the landmark that * is associated with must be fixed. To fix bad land marks lithos is used in the working directory.
How to Fix Landmarks
Once a problem landmark is identified, the executable lithos can be entered in the working directory to fix the landmark. The steps to do this are as follows:
Step 1
The first major step in fixing a bad landmark is to select that landmark within lithos. As an example, the landmark EM0004 in the find_nofit output above has major errors. To select this landmark the following commands are entered in the lithos executable:
i EM0004 n n
The output of these commands looks like this:
Current landmark = NONE
Main Menu: Q 0123O IUCRSG NVAMBX DEPL FZH? i
>i
Input 6-character landmark name.[Q]
EM0004
-0.31014 0.47683 -0.31113 0.29307
gc slope.pgm
Check for more images? y[n]
n
Include a single image? y[n]
n
Step 2
With a landmark identified in lithos, the next step is to align the landmark within various images of which it is present to one another. To do this, the following commands are typed into the lithos main menu.
1 0 1
The output of these commands looks like this.
Main Menu: Q 0123O IUCRSG NVAMBX DEPL FZH? 1
>1
0. Auto align
1. Individual shift
2. Global shift
3. Align with picture
4. Align with gradient
q. Quit
0
0
k picnm lambda phi cvg nlmk
1 M596105841F0 1.133 0.000 1.000 26
2 M596106238F0 1.271 0.000 1.000 26
3 S594918087F0 1.504 0.000 1.000 121
4 S594918496F0 1.078 0.000 1.000 120
5 S594918904F0 1.524 0.000 1.000 113
6 S594919721F0 0.984 0.000 1.000 81
7 S594920129F0 0.970 0.000 1.000 72
8 S594920537F0 1.602 0.000 1.000 67
9 S594920946F0 1.061 0.000 1.000 56
10 S594965887F0 1.521 0.000 1.000 85
11 S594966296F0 0.987 0.000 1.000 84
12 S594966704F0 0.946 0.000 1.000 80
13 S594967112F0 1.012 0.000 1.000 64
14 S594967521F0 1.015 0.000 1.000 54
15 S595057464F1 1.060 0.000 0.737 20
16 S595057569F1 1.011 0.000 0.998 45
17 S595057864F1 1.382 0.000 0.914 21
18 S595057964F1 1.370 0.000 1.000 58
19 S595058072F1 1.147 0.000 0.772 79
20 S595058359F1 1.033 0.000 1.000 24
21 S595058467F1 1.097 0.000 0.748 38
22 S595058649F1 1.446 0.000 0.575 28
23 S595058754F1 1.050 0.000 1.000 12
24 S595058862F1 1.443 0.000 0.566 31
25 S595059044F1 1.147 0.000 1.000 46
26 S595059154F1 1.086 0.000 0.871 16
27 S595059544F1 1.137 0.000 1.000 26
28 S595059939F1 1.581 0.000 1.000 19
29 S595060334F1 1.127 0.000 1.000 19
30 S595060837F1 1.732 0.000 1.000 14
31 S595152375F1 1.185 0.000 1.000 33
32 S595626396F2 1.629 0.000 0.708 18
33 S595626804F2 1.581 0.000 1.000 15
34 S595627212F2 1.373 0.000 1.000 14
35 S595627621F2 1.388 0.000 1.000 11
36 S595628029F2 1.322 0.000 0.585 12
37 S595707896F3 1.278 0.000 1.000 4
gc LMRK_DISPLAY0.pgm
gc LMRK_DISPLAY1.pgm
check display
enter spacing
1
1
1 M596105841F0 -0.073 -0.051 0.824 ++++
2 M596106238F0 0.116 -0.765 0.838 ++++
3 S594918087F0 -0.076 -0.082 0.757 ++++
4 S594918496F0 -0.054 -0.053 0.771 ++++
5 S594918904F0 -0.037 -0.030 0.831 ++++
6 S594919721F0 -0.015 -0.030 0.860 +++++
7 S594920129F0 -0.009 -0.018 0.852 ++++
8 S594920537F0 0.024 -0.009 0.874 +++++
9 S594920946F0 0.053 -0.014 0.853 ++++
10 S594965887F0 -0.016 -0.013 0.938 +++++
11 S594966296F0 -0.017 -0.009 0.919 +++++
12 S594966704F0 -0.009 -0.008 0.926 +++++
13 S594967112F0 0.008 -0.012 0.916 +++++
14 S594967521F0 0.030 0.003 0.919 +++++
15 S595057464F1 -0.111 -0.039 0.759 ++++
16 S595057569F1 -0.022 -0.026 0.824 ++++
17 S595057864F1 1.053 -0.614 0.713 +++
18 S595057964F1 -0.015 -0.013 0.894 +++++
19 S595058072F1 -0.032 -0.029 0.892 +++++
20 S595058359F1 -0.017 -0.013 0.905 +++++
21 S595058467F1 -0.013 -0.015 0.879 +++++
22 S595058649F1 0.021 0.038 0.809 ++++
23 S595058754F1 -0.014 -0.010 0.841 ++++
24 S595058862F1 -0.010 -0.009 0.940 +++++
25 S595059044F1 0.014 0.007 0.893 +++++
26 S595059154F1 -0.004 -0.011 0.932 +++++
27 S595059544F1 -0.010 -0.014 0.932 +++++
28 S595059939F1 0.011 0.006 0.920 +++++
29 S595060334F1 -0.001 -0.032 0.897 +++++
30 S595060837F1 0.030 0.029 0.888 +++++
31 S595152375F1 0.021 0.019 0.906 +++++
32 S595626396F2 -0.016 -0.013 0.766 ++++
33 S595626804F2 0.003 0.027 0.891 +++++
34 S595627212F2 -0.033 -0.043 0.838 ++++
35 S595627621F2 0.049 -0.067 0.859 +++++
36 S595628029F2 0.114 -0.155 0.781 ++++
37 S595707896F3 -0.037 -0.058 0.657 +++
0.177 0.156
new spacing? y[n]